One way of understanding the different conformations the ribosome can take on, depending on modification state or mRNA sequence, is to use Molecular Dynamics Markov State Modeling (MD-MSM). Markov State Models divide our MD trajectories into sets of different states based on atomic position. A key feature of MD-MSM is that each frame in the trajectory is treated as independent from every other frame, so the history of different states is assumed to be irrelevant to the current state. For this reason, it is important to use only the equilibrated portion of our trajectories in the analysis. K-means is the specific algorithm we use to separate the trajectories into different states, called clusters.
K-means Description
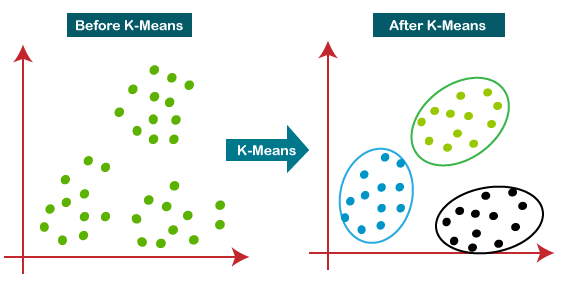
To understand how k-means works, imagine each green point in the figure above-left is the structure of the ribosome during one frame of the trajectory. The x- and y-axes capture multidimensional spatial information so that proximity of two points on the plot indicates similarity between two conformations of the ribosome. The goal of k-means is to assign each point to a cluster of similar structures, as indicated above-right.
This is done by selecting a range of k-values (number of clusters) to try. In the above example, k=3. In this case, three points are randomly selected to be the centroids of each cluster. All the other points are then assigned to the cluster with the most similar centroid by mathematically comparing atomic positions to find the minimum distance. The idea is for the centroid to be the best representative structure for all the other structures in a given cluster, but because the centroids are chosen randomly the first guess is expected to be quite bad. The solution is to iteratively adjust the chosen centroids, recalculate cluster membership, and evaluate whether or not those centroids are better (if the distances between cluster members and the centroid are lower). After many rounds of k-means, we accept the results and can learn about different states of the ribosome based on the centroids and cluster membership.
K-means Setup
1. Copy the entire k-means folder into your analysis directory.
2. Make a common topology (prmtop) file.
- The goal is to create a prmtop file that can be used for multiple experimental constructs (nucleotide substitutions, modifications, etc.). This can be done by reducing the prmtop to include only nucleotide and amino acid backbone atoms.
- We are starting with the version of the trajectories created in the data backup step (cpptraj 2), which have already been stripped of water and ions. Therefore we need the nowat version of the prmtop created during tLeAP. Choose one of the constructs you will be analyzing with k-means (it doesn't matter which) and copy its nowat.prmtop into CLUST_FILES.
- Modify k1_strip_prmtop.in and k1_strip_prmtop.sh as needed and run. This script will remove all non-backbone atoms from the entire ribosome prmtop.
3. Concatenate all 30 experiments together, repeat for each construct.
- The goal is to combine all 30 experiments together into one large trajectory that represents the equilibrium dynamics of one construct.
- Again, we are using the data backup trajectories, so you will need to run (cpptraj 2) at this point if you haven't already. These trajectories already have the first 20ns (non-equilibrium MD) removed and reduced to every 50th frame of the original trajectory (2 frames per ns remain).
- Modify k2_combine_30expts.py as needed and run for your first construct. Make sure you are pointing to the nowat.prmtop in your TLEAP folder (not the prmtop created in step 2) and the 1in50 trajectories in your backup folder. In addition to concatenation, this script strips out all non-backbone atoms.
- Repeat for all constructs in the k-means analysis. For example, if you are comparing A-CCU +1-GCU, A-CCU +1-CGU, A-CCC +1-GCU, and A-CCC +1-CGU, you will run the script 4 times.
4. Combine all constructs together into one trajectory.
- The goal is to combine all the experiments from all constructs to be analyzed together into one mega trajectory of equilibrium dynamics.
- Modify k3_combine_all_constructs.py as needed and run. Include all constructs created in step 3. Because the input trajectories from the previous step have non-backbone atoms and solvent removed, make sure you are pointing to the prmtop created in step 2.
- Note the order you are reading in the large trajectories. Knowing the number of frames (3200) and sequence of the constructs will allow you to analyze the k-means output in Excel later.
5. Make a common reference structure.
- The goal is to take the mega trajectory created in step 4 and create a structure that represents the average position of each atom across all frames. This structure will be used as the reference for an RMS fit, which will align each frame of the mega trajectory to the average structure as much as possible before running the k-means algorithm.
- Modify k4_make_reference.in and k4_make_reference.sh as needed and run. Because the backbone-only trajectory from step 4 is the input, make sure you are pointing to the prmtop created in step 2.
6. Run RMS fit and k-means.
- From within the CLUST_2 folder, modify kclust2.in and run_kmeans.sh as needed and run. Because the backbone-only reference structure from step 5 and the backbone-only trajectory from step 4 are the inputs, make sure you are pointing to the prmtop created in step 2.
- The RMS fit will align each frame of the trajectory to the reference as much as possible, relying only on backbone atoms.
- The k-means algorithm will be run on non-onion shell residues only, using only backbone atoms.
- Repeat for CLUST_3 to CLUST_8. You can have k=2 through k=7 running at the same time. Each k-means calculation will take about 30-60 minutes total (for 4 constructs, 12,800 frames).
7. Analyze the k-means results.
- See the K-means Analysis Tutorial below for detailed instructions.
Paths to Files
K-means Folder:
/home66/kscopino/AMBER22/BIN/KMEANS/
K-means Setup Tutorial (on Google Drive):
/ribosome/Molecular Dynamics/Tutorials and Starting Structures/Part4_Analysis_kmeans1_setup.mp4
K-means Analysis Tutorial (on Google Drive):
/ribosome/Molecular Dynamics/Tutorials and Starting Structures/Part4_Analysis_kmeans2_analysis.mp4