|
Background Workflow |
Site /
TranslationBiologyTranslation Overview The central dogma of biology is that DNA is transcribed into RNA by RNA polymerase, which is then translated into protein by the ribosome. The building blocks of RNA and protein are nucleotides and amino acids, respectively, and these building blocks are collectively referred to as "residues." RNA molecules can appear as several types of structures: mRNA (the template used in translation), tRNA (the molecules that recognize codons and are charged with the amino acids that are incorporated into the polypeptide chain), and rRNA (structural RNA of ribosomes). The ribosome is a large molecular machine that consists of several rRNA and protein chains. Nucleotides consist of a phosphate group, a sugar, and a base. The phosphate and sugar groups are linked together to form the sugar-phosphate backbone. There are four bases in RNA: adenine (A), guanine (G), cytosine (C), or uracil (U), each presenting an edge that canonically forms hydrogen-bonds (the Watson-Crick edge) and an edge that can form non-canonical hydrogen bonds (the Hoogstein edge). The sugar-edge can also participate in hydrogen bonding. Hydrogen bonding is one of the three types of reversible molecular interactions. The bases are categorized as pyrimidines (one ring - C, U) or purines (two rings - A, G), and this ring structure with delocalized pi electrons allows the bases to stack on top of each other. These delocalized electrons induce transient dipoles that produce attractive van der Waals forces, a second type of reversible molecular interaction.  Amino acids consist of a central alpha carbon which is bonded to an amino group, a carboxyl group (linkages of these three components make up the protein backbone), a hydrogen, and a characteristic R-group. The 20 different R-groups define each type of amino acid and its characteristics. R-groups vary in electrostatic character (charged, polar, or non-polar; the third type of reversible molecular interaction), structure (aliphatic or aromatic), size, shape, and hydrogen bond donor/acceptor pattern. Protein structure can be organized hierarchically into primary, secondary, tertiary, and quaternary structure. Primary structure is the sequence of amino acids. Secondary structure is when the primary sequence is organized in regular-repeating structures like alpha helices and beta strands, with loop regions in between. Tertiary structure is the low-energy, folded form that an amino-acid chain takes. Finally, quaternary structure is how multiple chains (subunits) come together to form multimers. Translation Ribosomes are responsible for translating mRNA into protein. Translation is initiated when a complex of eIF4E (5' cap-binding protein), a MET-tRNA, eIF1A (an initiation factor), and the small subunit scan the mRNA. eIF1A recognizes the AUG start codon, proteins are released from the complex, and the large subunit binds. Elongation occurs when amino acids are added to form a polypeptide chain. The residues of mRNA can be grouped into 3 nucleotide codons, where each codon specifies the amino acid to be incorporated next into the polypeptide chain. Codons are "read" by tRNA molecules which have a 3 nucleotide anticodon sequence that is complementary to the codon of the mRNA, and are bound to the amino acid specific to the codon. tRNAs enter the A-site of the ribosome, and base pairing of the codon and anticodon will occur if they are complementary. A bond will form between the last amino acid added to the chain and the amino acid attached to the tRNA. The ribosome will then ratchet over to the next codon in the mRNA 5' to 3' direction, moving the tRNA with the polypeptide chain attached into the P-site. The process repeats, and the tRNA in the P-site moves into the E-site where it exits the ribosome. This occurs until one of the three stop codons is reached, which results in translation termination. 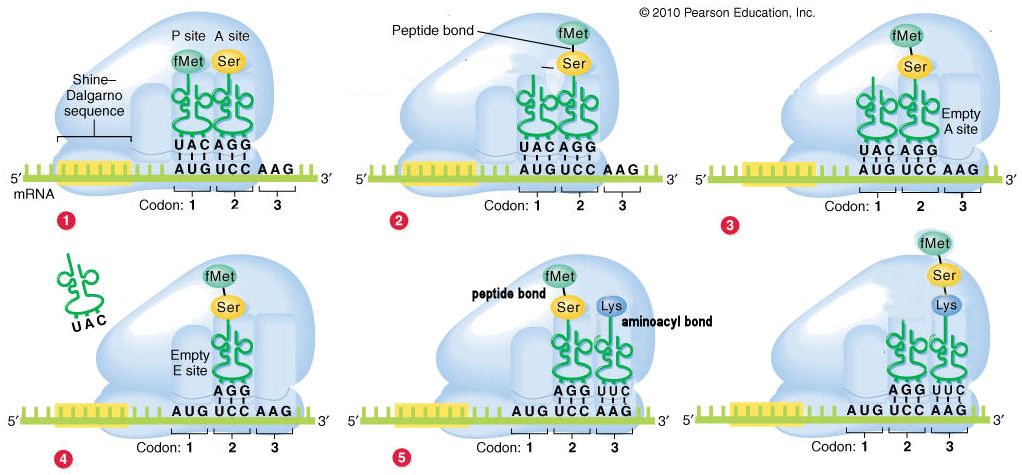 Ribosome Synthesis and Degradation Ribosome synthesis can be broken into four steps. The first is the transcription of rDNA into rRNA by RNA polymerase I. Next, covalent modification occurs. Primarily this is accomplished by snoRNAs, but some stand-alone enzymes are also responsible for modification. In the small subunit, this usually occurs co-transcriptionally while in the large unit, this can occur co-transcriptionally or post-transcriptionally. The main modifications to rRNA are ribose 2'-O-methylation and pseudouridylation. The methylation modifications may increase hydrophobicity and promote base-stacking, stabilizing helices (Liang, 2007). The pseudouridylations increase the hydrogen bond capability of uracil and increase the rigidity of the sugar-phosphate backbone. The transcript is then processed through cleavage by nucleases. Finally, the matured rRNA is assembled into the ribosomal subunits. |