|
Background Workflow |
Site /
T-LEaPThe first role of tLEaP is to add in any missing atoms based on a predefined standard geometry and parameterize all atoms based on the implemented force field (ff14SB for protein, OL3 for RNA, modrna08 for RNA modifications, and tip3p for water) and any experimental force field modifications. In addition to any experimental modifications our ribosome structures are complexed with the elongation factor eEF2, which contains the post-translationally modified histidine residue diphthamide. This residue is parameterized using the DDE modification files (see below). Our eventual goal is to determine the motion of atoms over time, i.e. perform molecular dynamics. The second role of tLEaP is to specify the details of the input to the equations used to calculate atomic motion. This first requires us to calculate (using tLEaP) the potential energy of each individual atom in the subsystem by using a multi-term equation with terms representing the bonds between atoms, the angles of those bonds, torsions, improper dihedrals, interactions between neutral atoms (using the Lennard-Jones potential), and interactions between charged atoms (using Coulomb's law). The negative derivative of this potential energy yields the force on an atom. Newton's second law is then used to calculate the atom's acceleration, adding the temporal component (during Heating, Equilibration, and Neutral Dynamics). 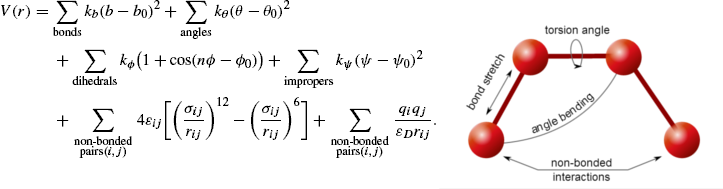 The third function of tLEaP is to define the system shape and solvate by specifying the box size and periodic boundary conditions, and adding water molecules and potassium counterions to make the overall system charge neutral. Amber18 tLEaP sample input # call Amber18 version of tLEaP # you will see a welcome message and new prompt icon > /share/apps/CENTOS7/amber/amber18/bin/tleap ### load in force field libraries ### # for protein source /home/apps/CENTOS7/amber/amber18/dat/leap/cmd/leaprc.protein.ff14SB # for RNA source /home/apps/CENTOS7/amber/amber18/dat/leap/cmd/leaprc.RNA.OL3 # for RNA modifications source /home/apps/CENTOS7/amber/amber18/dat/leap/cmd/leaprc.modrna08 # for water and ions source /home/apps/CENTOS7/amber/amber18/dat/leap/cmd/leaprc.water.tip3p ### load in force field modifications ### # needed for modified histidine, used for every project loadAmberPrep /home66/kscopino/FORCE_FIELDS/dde.prepin loadAmberParams /home66/kscopino/FORCE_FIELDS/frcmod1.dde loadAmberParams /home66/kscopino/FORCE_FIELDS/frcmod2.dde ### load any in project specific modification if needed (example: mR146) ### loadAmberPrep /home66/kscopino/FORCE_FIELDS/ffptm.in ### loadAmberParams /home66/kscopino/FORCE_FIELDS/DA1.frcmod # load in PDB x = loadpdb 5JUP_N2_GCU_pretleap.pdb # check for errors, charge and close contact warnings are okay check x # save stripped version saveAmberParm x 5JUP_N2_GCU_nowat.prmtop 5JUP_N2_GCU_nowat.rst # add potassium counterions to achieve overall electrical neutrality addions x K+ 0 # add water in an octahedral box with 12 angstrom periodic boundaries using the TIP3P explicit solvent model # this distance allows for the presence of two layers of solvent shell solvateOct x TIP3PBOX 12.0 # check for errors, close contact warnings are okay check x # save solvated version saveAmberParm x 5JUP_N2_GCU_wat.prmtop 5JUP_N2_GCU_wat.rst quit Amber22 tLEaP sample input # call Amber22 version of tLEaP # you will see a welcome message and new prompt icon > /share/apps/CENTOS7/amber/amber22/bin/tleap ### load in force field libraries ### # for protein source /home/apps/CENTOS7/amber/amber22/dat/leap/cmd/leaprc.protein.ff14SB # for RNA source /home/apps/CENTOS7/amber/amber22/dat/leap/cmd/leaprc.RNA.OL3 # for RNA modifications source /home/apps/CENTOS7/amber/amber22/dat/leap/cmd/leaprc.modrna08 # for water and ions source /home/apps/CENTOS7/amber/amber22/dat/leap/cmd/leaprc.water.tip3p ### load in force field modifications ### # needed for modified histidine, used for every project loadAmberPrep /home66/kscopino/FORCE_FIELDS/dde.prepin loadAmberParams /home66/kscopino/FORCE_FIELDS/frcmod1.dde loadAmberParams /home66/kscopino/FORCE_FIELDS/frcmod2.dde ### load any in project specific modification if needed (example: mR146) ### loadAmberPrep /home66/kscopino/FORCE_FIELDS/ffptm.in ### loadAmberParams /home66/kscopino/FORCE_FIELDS/DA1.frcmod # load in PDB x = loadpdb 5JUP_N2_GCU_pretleap.pdb # check for errors, charge and close contact warnings are okay check x # save stripped version saveAmberParm x 5JUP_N2_GCU_nowat.prmtop 5JUP_N2_GCU_nowat.rst # add potassium counterions to achieve overall electrical neutrality addions x K+ 0 # add water in an octahedral box with 12 angstrom periodic boundaries using the TIP3P explicit solvent model # this distance allows for the presence of two layers of solvent shell solvateOct x TIP3PBOX 12.0 # check for errors, close contact warnings are okay check x # save solvated version saveAmberParm x 5JUP_N2_GCU_wat.prmtop 5JUP_N2_GCU_wat.rst quit tLEaP output x_nowat.rst -- Stripped (no water or ions) single frame containing atomic coordinates x_nowat.prmtop -- Stripped (no water or ions) parameter file containing force field information x_wat.rst -- Solvated (contains water and ions) single frame containing atomic coordinates, used as starting structure for energy minimization x_wat.prmtop -- Solvated (contains water and ions) parameter file containing force field information, needed for energy minimization calculations At this point it is helpful to use cpptraj to create a PDB from your tLEaP output to check overall system appearance and make sure any intended modifications occurred (path below). Because the connectivity information is inferred in PyMOL from the PDB, atoms may appear bonded when they are close together. Paths to FilesRST to PDB Script: /home66/kscopino/AMBER18/BIN/make_pdb_from_rst.in /home66/kscopino/AMBER22/BIN/make_pdb_from_rst.in cpptraj Job Submission Script: /home66/kscopino/AMBER18/BIN/run_cpptraj.sh /home66/kscopino/AMBER22/BIN/run_cpptraj.sh Tutorials (on Google Drive): /ribosome/Molecular Dynamics/Tutorials and Starting Structures/Part1_tLEaP.mp4 WITH CORRECTION: /ribosome/Molecular Dynamics/Tutorials and Starting Structures/Part1_tLEaP_correction.mp4 |